RECOT: Read Coordinate Transformer
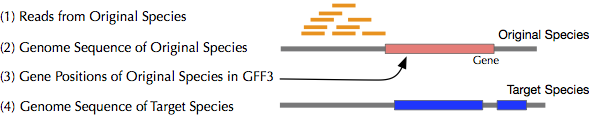
RECOT transforms coordinate of next-generation sequencer reads from your genome/gene to other species genome. RECOT is useful to compare RNA-seq, ChIP-seq and CLIP-seq sequences between closely-related species.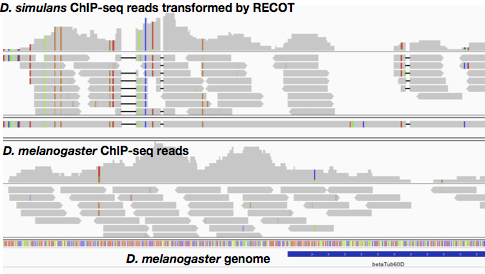
Requirements (Data)
- Reads of your original species (1)
- Two different genomes (Your originally experimented species and target species) (2,4)
- Gene regions of your original genome in GFF format (3)
- (Option) Relationships of genes between two species
Requirements (Software)
- Python (>=2.6)
- Short read alignment program, such as BWA and Bowtie
- Alignment program for long sequence, such as GMAP/GSNAP
- Both types of alignment programs must support SAM output because our programs use the format
Simple Usage
As example, we here transform coordinate of ChIP-seq reads on D. simulans to D. melanogaster genome. "sample" directory contains chromosome 2R of both D. simulans and D. melanogaster, and SAM file contains ChIP-seq result mapped onto D. simulans genome. Scripts will suggest you the next action. By changing settings in configuration file "settings.ini", you can change both input and output file names.
Install [First time only]
If you have Git, you can clone latest version by running:
git clone git://github.com/sesejun/recotDon't worry when you do not have Git! Download the zipped codes and extract it.
Extract gene sequences
First, extract gene sequences. "recot_extract.py" will generate "sample/dsim_geneseq.fasta" which contains gene and its regulatory regions (upstream and downstream 500bp as default).
python recot_extract.py -c settings_sample.ini

Align the gene sequences against target genome
Map the sequences in "data/dsim_geneseq.fasta" onto target genome. We here use GMAP to map the sequences.
gmap_build -d dme sample/dmel-2R.fasta gmap -f samse -d dme sample/dsim_geneseq.fasta > sample/dsim_geneseq_on_dme.samBy changing gmap option "samse" to "sampe", it can handle paired end sequence. Then, you can select the best matched sequences from the SAM file.
python recot_rm_overlap.py -c settings_sample.ini

Map short reads against target genes/genome
Map your reads onto original genome sequence (In the running example, D. simulans). Mapped result file in this sample is "data/ERR020078-2R-1M.sam", which only contains reads mapped onto the beginnings of 1Mbp of chromosome 2R.
Change coordinate of reads according to the gene sequence mapped result
Combine gene sequence mapped result against reference sequence and read mapped result against target genes/genomes.
python recot_combine.py -c settings_sample.ini python recot_convert.py -c settings_sample.iniThis step may require a few hours (depend on number of reads and CPU/Disk speed).

Visualize the result
When recot_convert.py is successfully finished, the script shows the way to convert the result SAM file into BAM file.
samtools view -bt sample/dmel-2R.fasta -o your_read.bam sample/ERR020078.on_dme.sam samtools sort your_read.bam sample/ERR020078.on_dme.bam samtools index sample/ERR020078.on_dme.bamTo visualize the result in IGV, you can use the ERR020078.on_dme.bam (and ERR020078.on_dme.bam.bai).
FAQ
Q. Can I change configuration file?
By adding option "-c", you can change the file name from "settings.ini".
Q. Can I transform coordinate of RNA-seq reads even when I do not have genome sequence of original species?
Yes. First, you prepare gene sequences in FASTA format (From RNA-seq reads, you can use transcriptome assembly software such as Trinity and OASIS). Then, by skipping "recot_extract.py" in the above procedure and specify the result file at gene_seq_file parameter in "setting.ini", you can transform coordinate of the reads.
Q. I would like to use prepared orthologous gene relationships between two species.
recot_rm_overlap.py can handle the orthologous genes. To use the relationships,
- Prepare the tab delimited file containing gene-gene relation (sample/relation_dsim_dme.tsv). First column is gene name of original species, and second column is gene name of target species.
- In setting.ini, you need to change two parameters. 1. change the value of "gene_set" to the file including orthologous genes. 2. set target_gff_file, which contains gene names and their regions of target species with GFF format.
Q. Samtools give up convert of SAM to BAM halfway with some errors. What's wrong?
Bugs in mapping software or our scripts sometime cause the problem. In the case, please run the following command and then use the "output.sam" instead of input.sam:
python recot_check_cigar.py input.sam output.sam errorreads.sam
Q. I have billions of reads. Can I increase the speed of the transformation?
If you can use SUN(Oracle) Grid Engine or Torque, you can parallelize the transformation in "recot_convert.py" by using array job. First, you need to count the number of chromosomes (contigs) of your original species (Suppose that we have 20 contigs). Then, run the following command instead of running "recot_convert.py" (for Sun Grid Engine):
qsub -t1-20:1 qsub_convert.sh python recot_samjoin.pyTo change a setting file, you need to add "-c" in the command in qsub_convert.sh and run recot_samjoin.py with -c option.
License
BSD License
